KNN and K-means clustering
Gerko Vink
Erik-Jan van Kesteren
Methodology & Statistics @ Utrecht University
December 9, 2022
Packages and functions that we use
Custom theme for plots
Supervised learning
We want to find the predictive function:
\[Y = f(X) + \epsilon \]
That minimizes \(\epsilon\) with respect to our goal.
- Function \(f\) is an unknown, but fixed function of \(X = X1, \dots, Xp\)
- \(p\) is the number of predictors
- \(Y\) is the quantitative response
- \(\epsilon \sim N(0, \sigma_\epsilon^2)\) is a random error term
- \(\epsilon\) does not depend on \(X\)
Our aim is to find the \(f(X)\) that best represent the systematic information that \(X\) yields about \(Y\).
Supervised learning
With supervised learning every observation on our predictor
\[x_i, i=1, \dots, n\]
has a corresponding outcome measurement
\[y_i\] such that
\[\hat{y_i}=f({\bf x_i})\quad \text{and} \quad y_i = f({\bf x_i})+\epsilon_i.\]
Examples:
- linear regression
- logistic regression
- k-nearest neighbours
Unsupervised learning
With unsupervised learning we have a vector of measurement \(\bf x_i\) for every unit \(i=1, \dots, n\), but we miss the associated response \(y_i\).
There is no outcome to predict
- Hence you cannot fit e.g. a linear regression model
There is no outcome to verify the model
- We lack the truth to supervise our analysis
What can we do?
Find patterns in \(\bf x_1, \dots, x_n\)
We can use this model to e.g. find out if some cases are more similar than other cases or which variables explain most of the variation
Examples:
- Principal Components Analysis
- k-means clustering
K-means and K-nearest neighbours
Two nonparametric algorithms
K-nearest neighbours (KNN)
- supervised learning
- prediction
- classification
K-means clustering (kmeans)
- unsupervised learning
- dimension reduction / pattern recognition
- clustering
Example dataset
Let’s create some data from a multivariate normal distribution
We start with fixing the random seed
and specifying the variance covariance matrix:
Data relations
Because the variances are 1, the resulting data will have a correlation of \[\rho = \frac{\text{cov}(y, x)}{\sigma_y\sigma_x} = \frac{.5}{1\times1} = .5.\]
Let’s draw the data
Plot the data
Now add some clustering
We have added a new column that randomly assigns rows to level A, B or C
Plot the data again
Adjust the clusters to make them distinct
The result: supervised
The result: unsupervised
K-Nearest Neighbors
How does it work?
For every test observation \(x_0\) the \(K\) points that are close to \(x_0\) are identified.
These closest points form set \(\mathcal{N}_0\).
We estimate the probability for \(x_0\) being part of class \(j\) as the fraction of points in \(\mathcal{N}_0\) for whom the response equals \(j\): \[P(Y = j | X = x_0) = \frac{1}{K}\sum_{i\in\mathcal{N}_0}I(y_i=j)\]
The observation \(x_0\) is classified to the class with the largest probability
In short
An observation gets that class assigned to which most of its \(K\) neighbours belong
Why KNN?
Because \(X\) is assigned to the class to which most of the observations belong it is
non-parametric
- no assumptions about the distributions, or the shape of the decision boundary
expected to be far better than logistic regression when decision boundaries are non-linear
However, we do not get parameters as with LDA and regression.
- We thus cannot determine the relative importance of predictors
- The “model” == the existing observations: instance-based learning
Fitting a K-NN model
First we need to determine a training set
set.seed(123)
sim.data <-
sim.data %>%
mutate(set = sample(c("Train", "Test"), size = 100,
prob = c(.25, .75), replace = TRUE))
sim.data# A tibble: 100 × 4
x1 x2 class set
<dbl> <dbl> <chr> <chr>
1 5.90 6.13 C Test
2 8.02 6.97 C Train
3 7.07 8.19 C Test
4 3.62 5.40 A Train
5 2.72 5.89 A Train
6 7.78 7.16 C Test
7 5.42 3.71 B Test
8 6.43 8.08 C Train
9 4.97 1.73 B Test
10 4.06 6.22 A Test
# … with 90 more rowsFitting a K-NN model
Then we split the data into a training (build the model) and a test (verify the model) set
Now we can fit the K-NN model
Predictions
The (in)correct responses KNN = 3
Fewer neighbours
More neighbours
Even more neighbours
The (in)correct responses KNN = 10
Predicting a new observation
Let’s make a new observation:
Predicting a new observation
Predicting a new observation
Now we predict the class of this new observation, using the entire data for training our model
K-means clustering
Remember: unsupervised
Goal: finding clusters in our data
K-means clustering partitions our dataset into \(K\) distinct, non-overlapping clusters or subgroups.
What is a cluster?
A set of relatively similar observations.
What is “relatively similar”?
This is up to the programmer/researcher to decide. For example, we can say the “within-class” variance is as small as possible and the between-class variance as large as possible.
Why perform clustering?
We expect clusters in our data, but weren’t able to measure them
- potential new subtypes of cancer tissue
We want to summarise features into a categorical variable to use in further decisions/analysis
- subgrouping people by their spending types
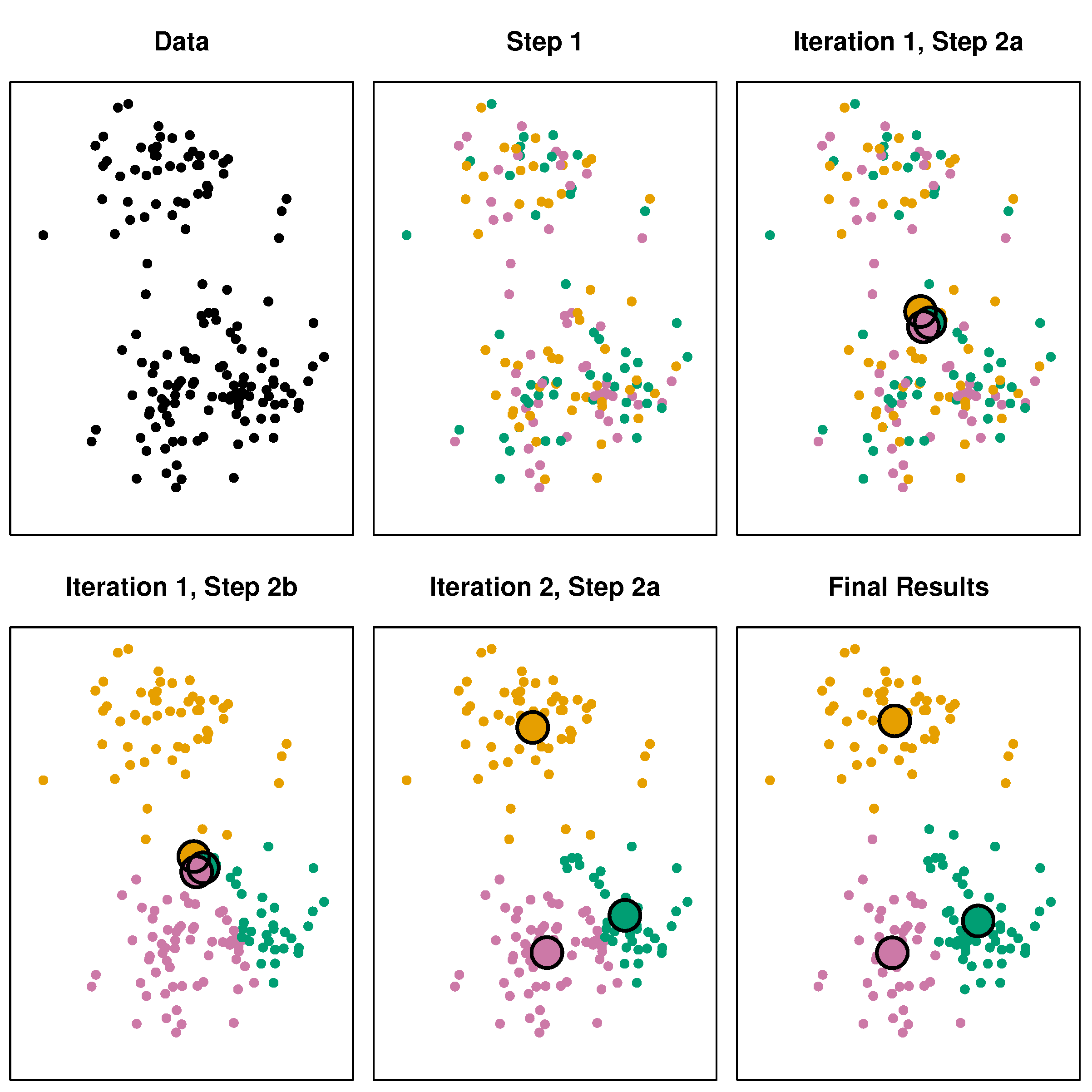
The k-means algorithm
- Randomly assign values to K classes
- Calculate the centroid (
colMeans) for each class - Assign each value to its closest centroid class
- If the assignments changed, go to step 2. else stop.
Source: James, G., Witten, D., Hastie, T., & Tibshirani, R. (2013). An introduction to statistical learning (Vol. 112). New York: Springer.
The k-means algorithm
K is a tuning parameter (centers)
K-means clustering with 3 clusters of sizes 39, 35, 26
Cluster means:
x1 x2
1 4.994727 3.532014
2 3.591445 6.394369
3 6.845077 6.976240
Clustering vector:
[1] 3 3 3 2 2 3 1 3 1 2 1 1 1 2 2 2 3 3 2 2 3 2 3 1 2 2 3 1 2 1 2 1 3 1 3 1 2
[38] 1 2 1 1 1 1 2 1 2 1 2 3 1 2 3 1 1 1 3 2 2 1 1 2 2 3 1 3 1 1 1 2 1 2 2 2 1
[75] 1 3 3 2 1 1 3 3 3 3 1 2 2 1 2 1 1 3 1 2 3 2 2 3 1 2
Within cluster sum of squares by cluster:
[1] 49.33578 46.51667 49.21032
(between_SS / total_SS = 73.0 %)
Available components:
[1] "cluster" "centers" "totss" "withinss" "tot.withinss"
[6] "betweenss" "size" "iter" "ifault" The result:
Comparison
Centroids
K = 5
K = 2

Gerko Vink