Statistics, Pipes and Visualization
12/9/22
Materials
All materials can be found at
www.gerkovink.com/rijkR
Disclaimer
I owe a debt of gratitude to many people as the thoughts and teachings in my slides are the process of years-long development cycles and discussions with my team, friends, colleagues and peers. When someone has contributed to the content of the slides, I have credited their authorship.
When external figures and other sources are shown:
- the references are included when the origin is known, or
- the objects are directly linked from within the public domain and the source can be obtained by right-clicking the objects.
Scientific references are in the footer.
Opinions are my own.
Packages used:
Vocabulary
Terms I may use
- TDGM: True data generating model
- DGP: Data generating process, closely related to the TDGM, but with all the wacky additional uncertainty
- Truth: The comparative truth that we are interested in
- Bias: The distance to the comparative truth
- Variance: When not everything is the same
- Estimate: Something that we calculate or guess
- Estimand: The thing we aim to estimate and guess
- Population: That larger entity without sampling variance
- Sample: The smaller thing with sampling variance
- Incomplete: There exists a more complete version, but we don’t have it
- Observed: What we have
- Unobserved: What we would also like to have
Statistical inference
At the start
We begin today with an exploration into statistical inference.
Truths are boring, but they are convenient.
- however, for most problems truths require a lot of calculations, tallying or a complete census.
- therefore, a proxy of the truth is in most cases sufficient
- An example for such a proxy is a sample
- Samples are widely used and have been for a long time1
Being wrong about the truth
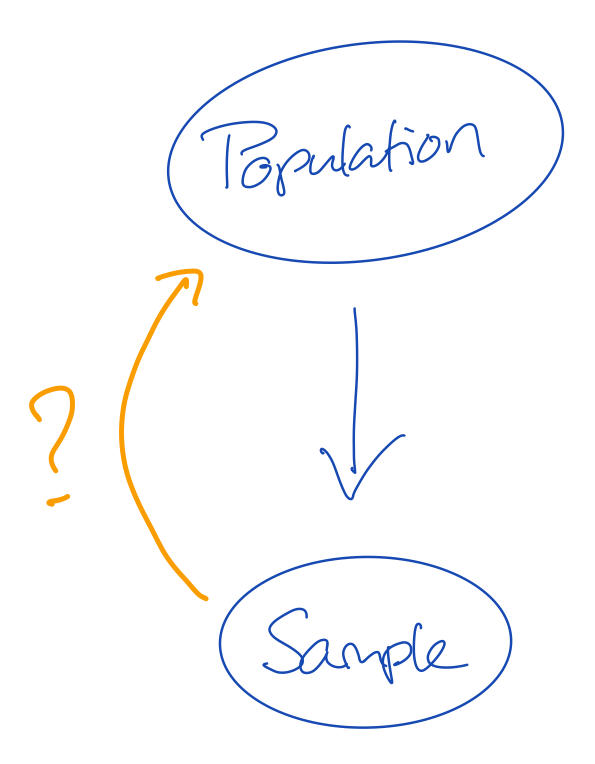
- The population is the truth
- The sample comes from the population, but is generally smaller in size
- This means that not all cases from the population can be in our sample
- If not all information from the population is in the sample, then our sample may be wrong
Q1: Why is it important that our sample is not wrong?
Q2: How do we know that our sample is not wrong?
Solving the missingness problem
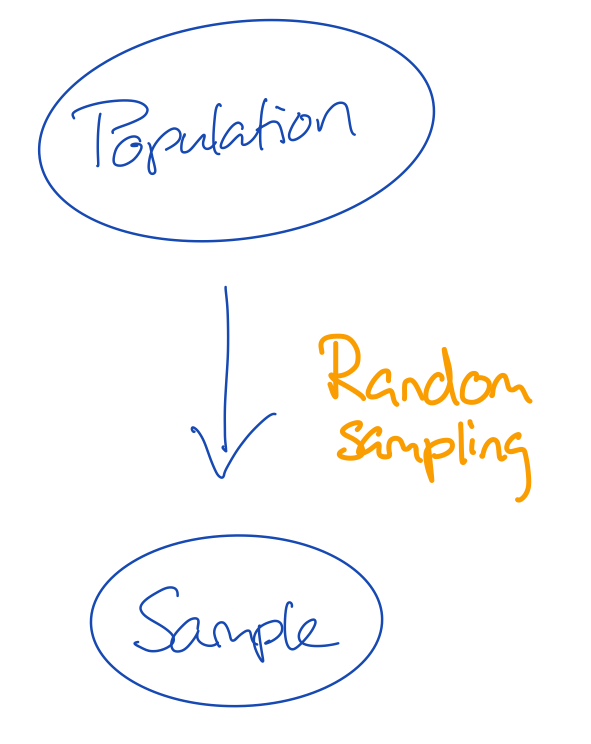
- There are many flavours of sampling
- If we give every unit in the population the same probability to be sampled, we do random sampling
- The convenience with random sampling is that the missingness problem can be ignored
- The missingness problem would in this case be: not every unit in the population has been observed in the sample
Q3: Would that mean that if we simply observe every potential unit, we would be unbiased about the truth?
Sidestep
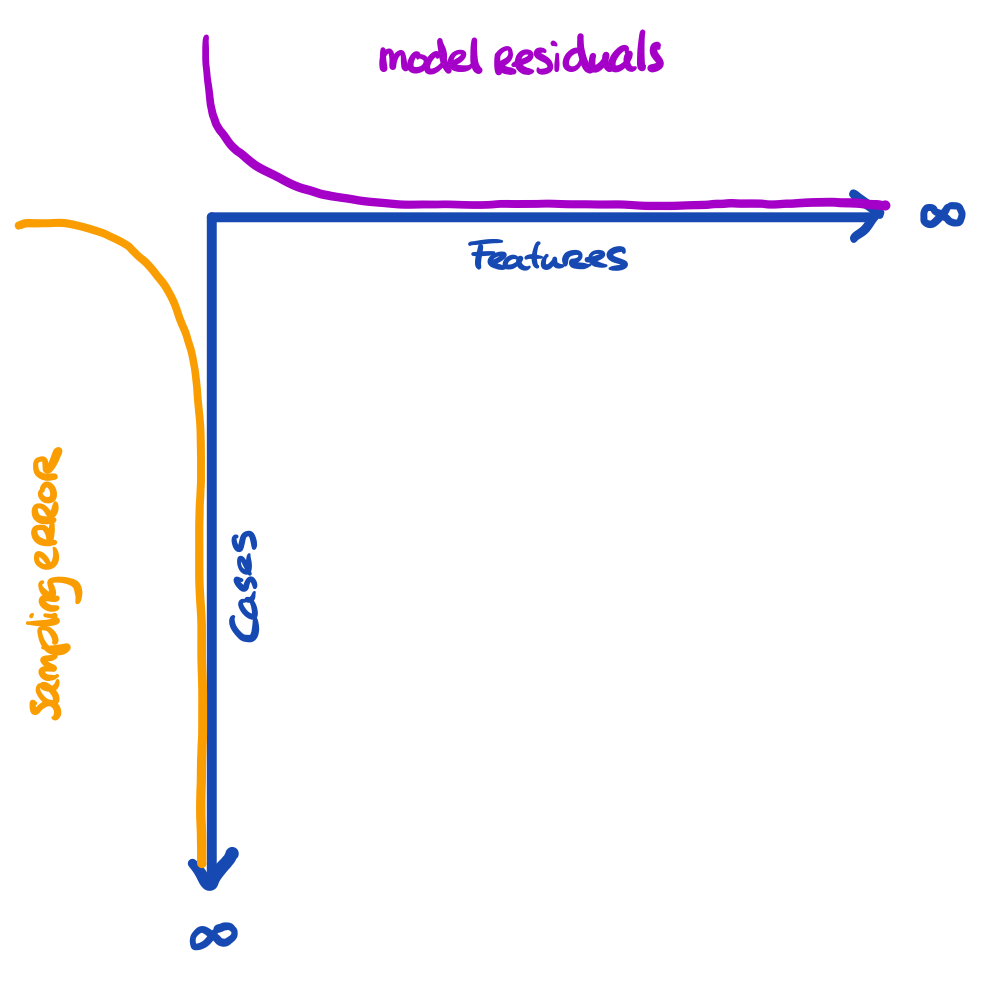
The problem is a bit larger
We have three entities at play, here:
- The truth we’re interested in
- The proxy that we have (e.g. sample)
- The model that we’re running
The more features we use, the more we capture about the outcome for the cases in the data
Sidestep

- The more cases we have, the more we approach the true information
All these things are related to uncertainty. Our model can still yield biased results when fitted to \(\infty\) features. Our inference can still be wrong when obtained on \(\infty\) cases.
Sidestep
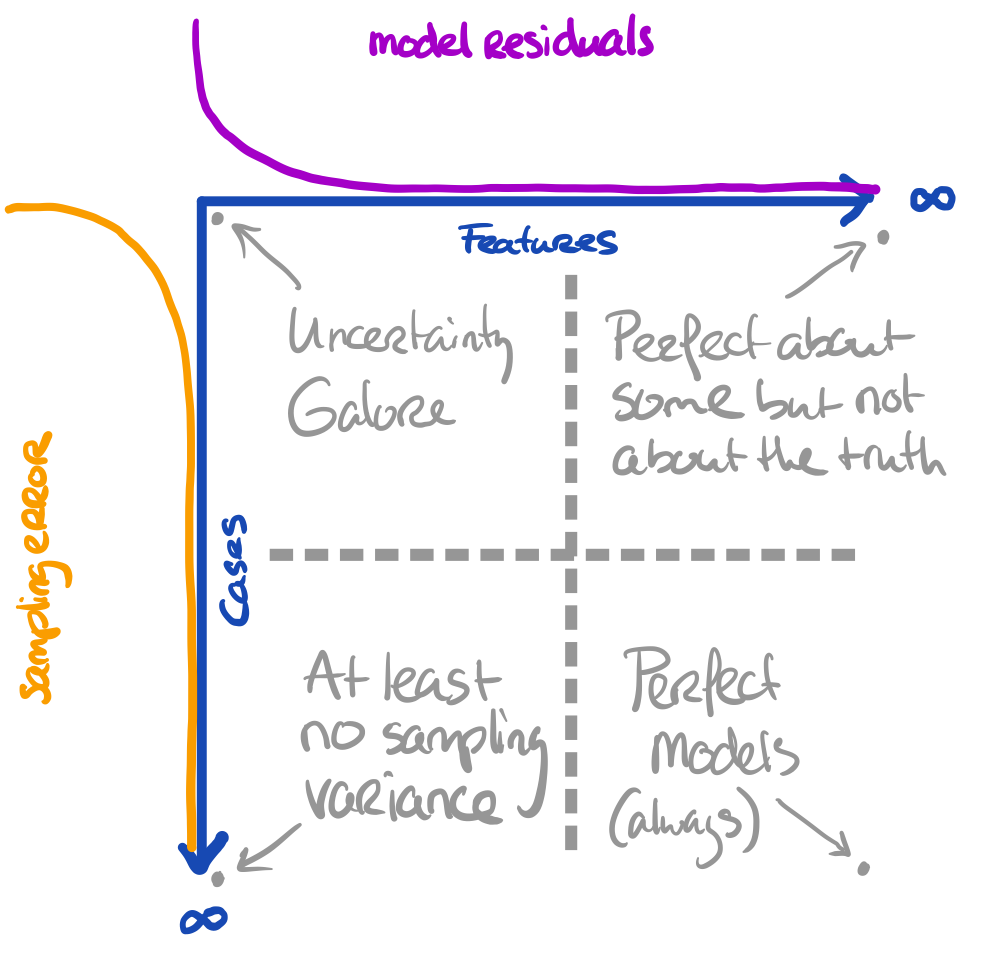
Core assumption: all observations are bonafide
Let’s start with R
Packages we use in these slides
The data
age hgt wgt bmi hc gen phb tv reg
3 0.035 50.1 3.650 14.54 33.7 <NA> <NA> NA south
4 0.038 53.5 3.370 11.77 35.0 <NA> <NA> NA south
18 0.057 50.0 3.140 12.56 35.2 <NA> <NA> NA south
23 0.060 54.5 4.270 14.37 36.7 <NA> <NA> NA south
28 0.062 57.5 5.030 15.21 37.3 <NA> <NA> NA south
36 0.068 55.5 4.655 15.11 37.0 <NA> <NA> NA southGoal
At the end of this lecture we aim to understand what happens in
Pipes
This is a pipe:
It effectively replaces round(cor(select(boys, is.numeric), use = "pairwise.complete.obs"), digits = 3).

Why are pipes useful?
Benefit: a single object in memory that is easy to interpret Your code becomes more readable:
- data operations are structured from left-to-right and not from in-to-out
- nested function calls are avoided
- local variables and copied objects are avoided
- easy to add steps in the sequence
What do pipes do:
f(x)becomesx %>% f()
f(x, y)becomesx %>% f(y)
age hgt wgt bmi hc gen phb tv reg
3 0.035 50.1 3.65 14.54 33.7 <NA> <NA> NA southh(g(f(x)))becomesx %>% f %>% g %>% h
More pipe stuff
The standard %>% pipe

The %$% pipe
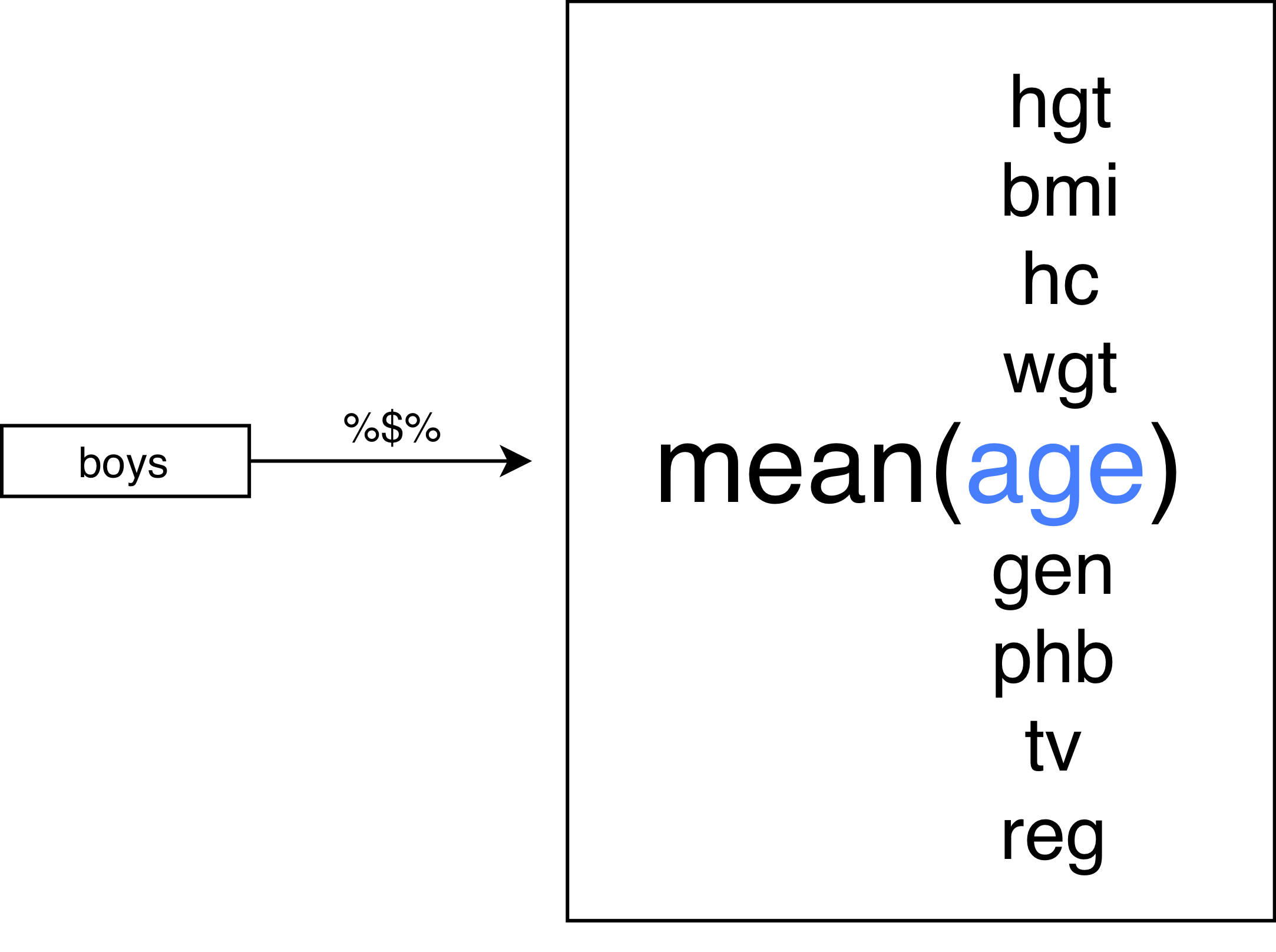
The role of . in a pipe
In a %>% b(arg1, arg2, arg3), a will become arg1. With . we can change this.
VS
The . can be used as a placeholder in the pipe.
Data manipulation
Performing a t-test in a pipe
Welch Two Sample t-test
data: age by ovwgt
t = -15.971, df = 32.993, p-value < 2.2e-16
alternative hypothesis: true difference in means between group FALSE and group TRUE is not equal to 0
95 percent confidence interval:
-9.393179 -7.270438
sample estimates:
mean in group FALSE mean in group TRUE
9.103392 17.435200 is the same as
Melting
Calculate statistics
Multiple columns
Mutate: add
Mutate: remove
Mutate: change
age hgt wgt bmi hc gen phb tv reg
7410 20.372 1.887 59.8 16.79 55.2 <NA> <NA> NA west
7418 20.429 1.811 67.2 20.48 56.6 <NA> <NA> NA north
7444 20.761 1.891 88.0 24.60 NA <NA> <NA> NA west
7447 20.780 1.935 75.4 20.13 NA <NA> <NA> NA west
7451 20.813 1.890 78.0 21.83 59.9 <NA> <NA> NA north
7475 21.177 1.818 76.5 23.14 NA <NA> <NA> NA eastMutate: transform column
Data visualization with ggplot2
The anscombe data
x1 x2 x3 x4 y1 y2 y3 y4
1 10 10 10 8 8.04 9.14 7.46 6.58
2 8 8 8 8 6.95 8.14 6.77 5.76
3 13 13 13 8 7.58 8.74 12.74 7.71
4 9 9 9 8 8.81 8.77 7.11 8.84
5 11 11 11 8 8.33 9.26 7.81 8.47
6 14 14 14 8 9.96 8.10 8.84 7.04
7 6 6 6 8 7.24 6.13 6.08 5.25
8 4 4 4 19 4.26 3.10 5.39 12.50
9 12 12 12 8 10.84 9.13 8.15 5.56
10 7 7 7 8 4.82 7.26 6.42 7.91
11 5 5 5 8 5.68 4.74 5.73 6.89The same statistical properties
x1 x2 x3 x4 y1 y2 y3 y4
9.000000 9.000000 9.000000 9.000000 7.500909 7.500909 7.500000 7.500909 y1 y2 y3 y4
x1 0.816 0.816 0.816 -0.314
x2 0.816 0.816 0.816 -0.314
x3 0.816 0.816 0.816 -0.314
x4 -0.529 -0.718 -0.345 0.817 y1 y2 y3 y4
x1 5.501 5.500 5.497 -2.115
x2 5.501 5.500 5.497 -2.115
x3 5.501 5.500 5.497 -2.115
x4 -3.565 -4.841 -2.321 5.499Fitting a line
Why visualise?
- We can process a lot of information quickly with our eyes
- Plots give us information about
- Distribution / shape
- Irregularities
- Assumptions
- Intuitions
- Summary statistics, correlations, parameters, model tests, p-values do not tell the whole story
ALWAYS plot your data!
Why visualise?
Anscombe, F. J. (1973). “Graphs in Statistical Analysis”. American Statistician. 27 (1): 17–21.
Why visualise?

What is ggplot2?
Layered plotting based on the book The Grammer of Graphics by Leland Wilkinson.
Wilkinson, L. (2006). The Grammar of Graphics. Springer Science & Business Media.
With ggplot2 you
- provide the data
- define how to map variables to aesthetics
- state which geometric object to display
- (optional) edit the overall theme of the plot
ggplot2 then takes care of the details
An example: scatterplot
1: Provide the data
2: map variable to aesthetics
3: state which geometric object to display
An example: scatterplot
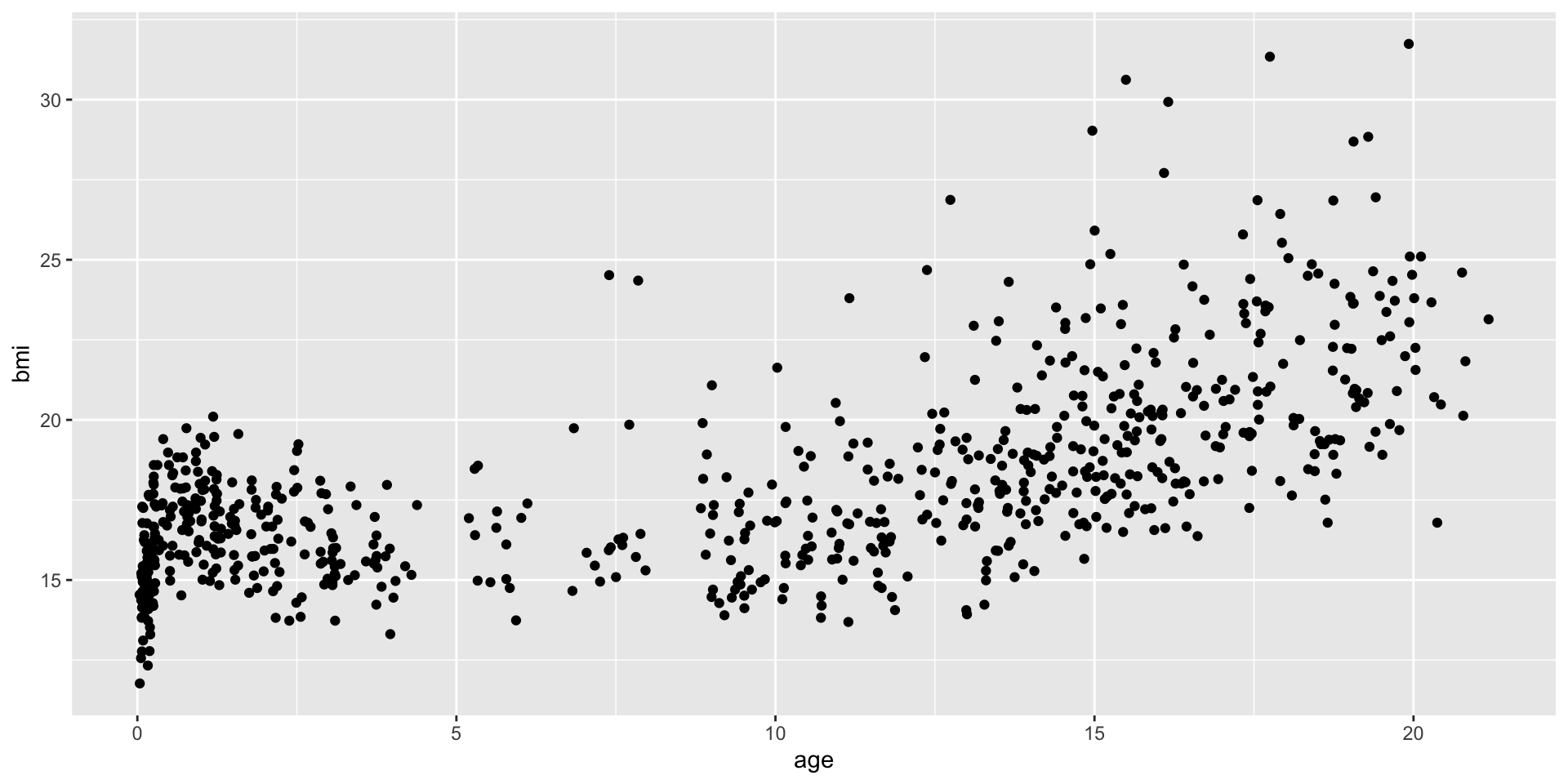
Why this syntax?
Create the plot
Add another layer (smooth fit line)
Give it some labels and a nice look
Why this syntax?
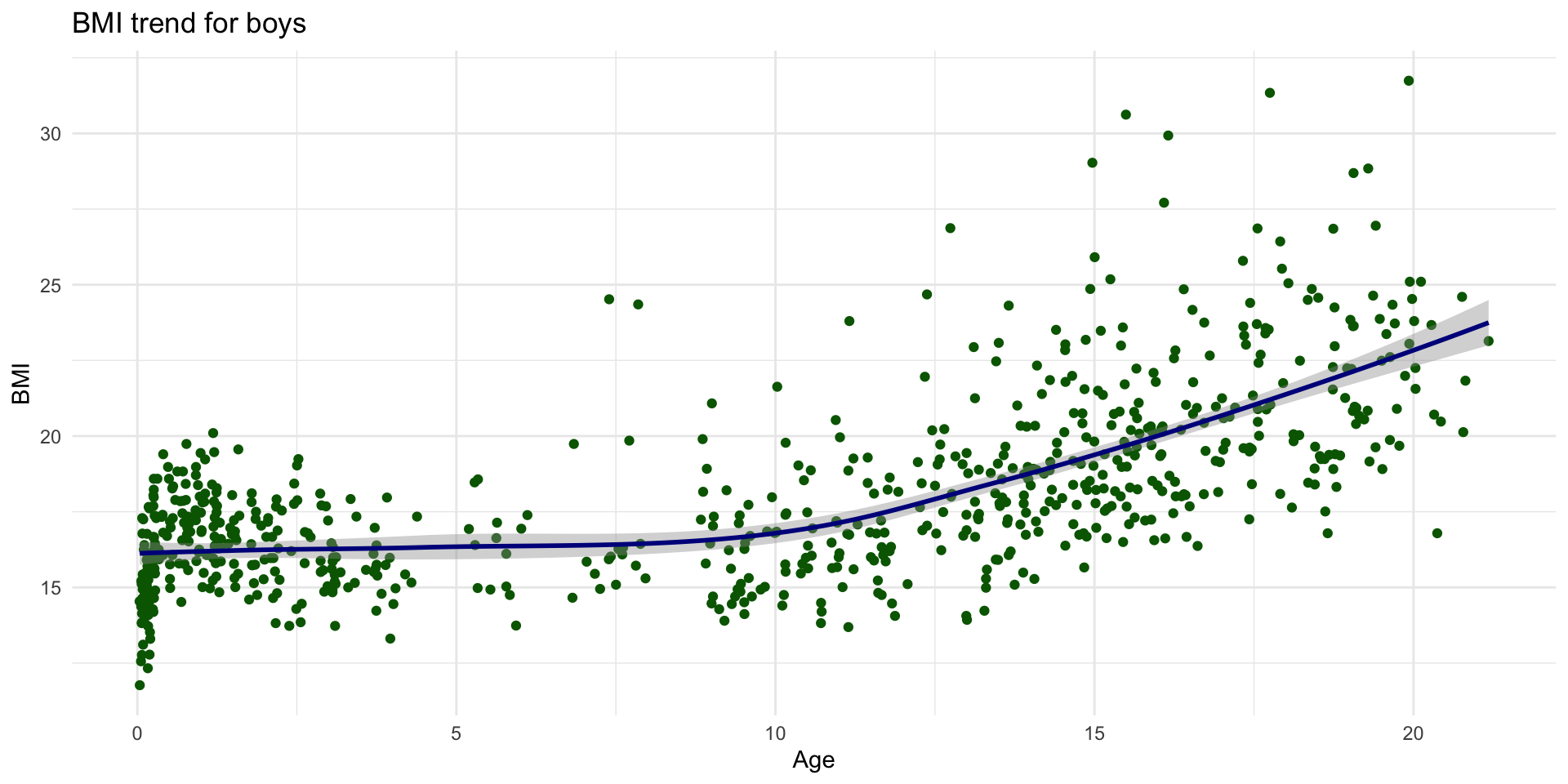
Why this syntax?
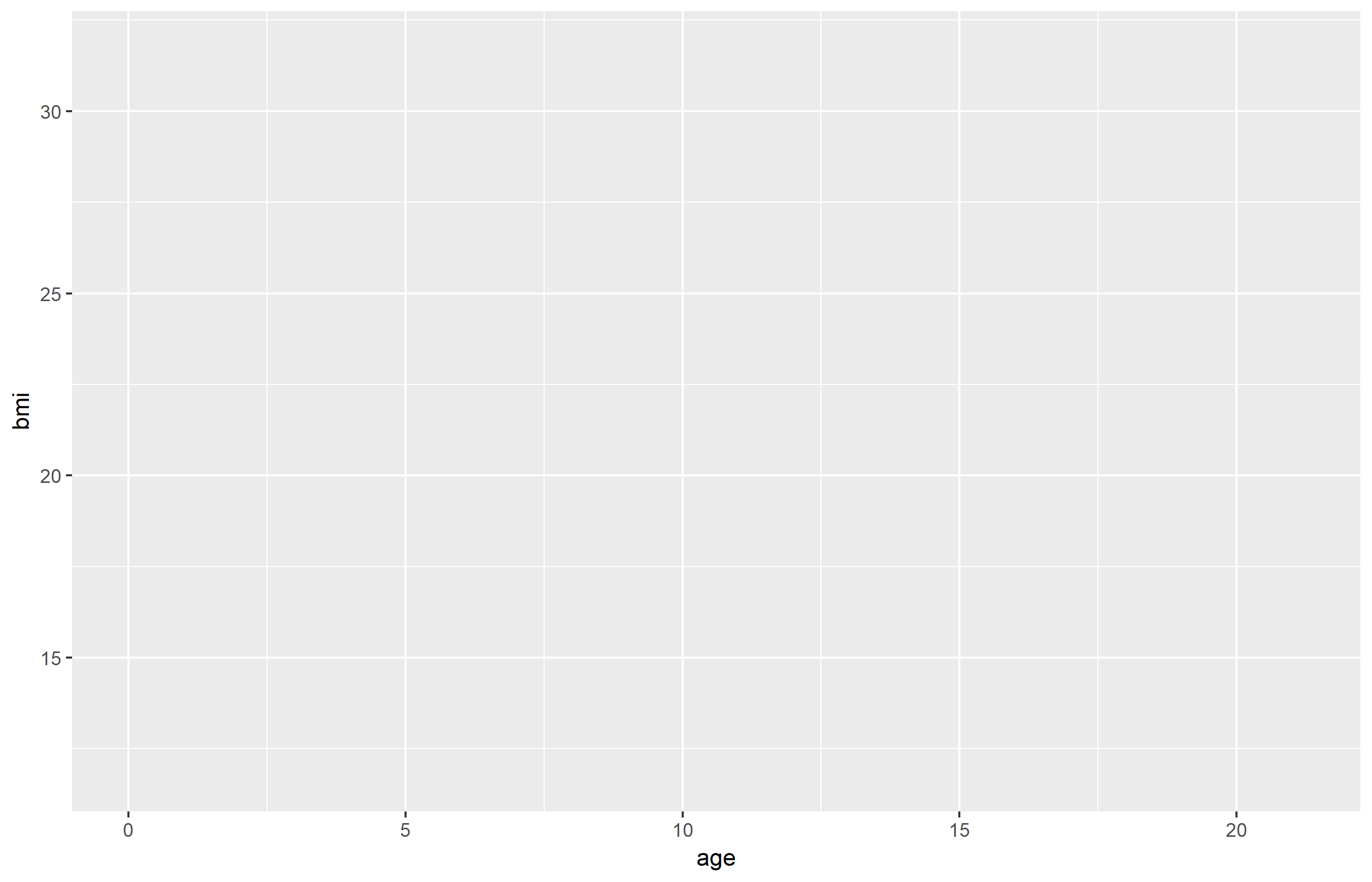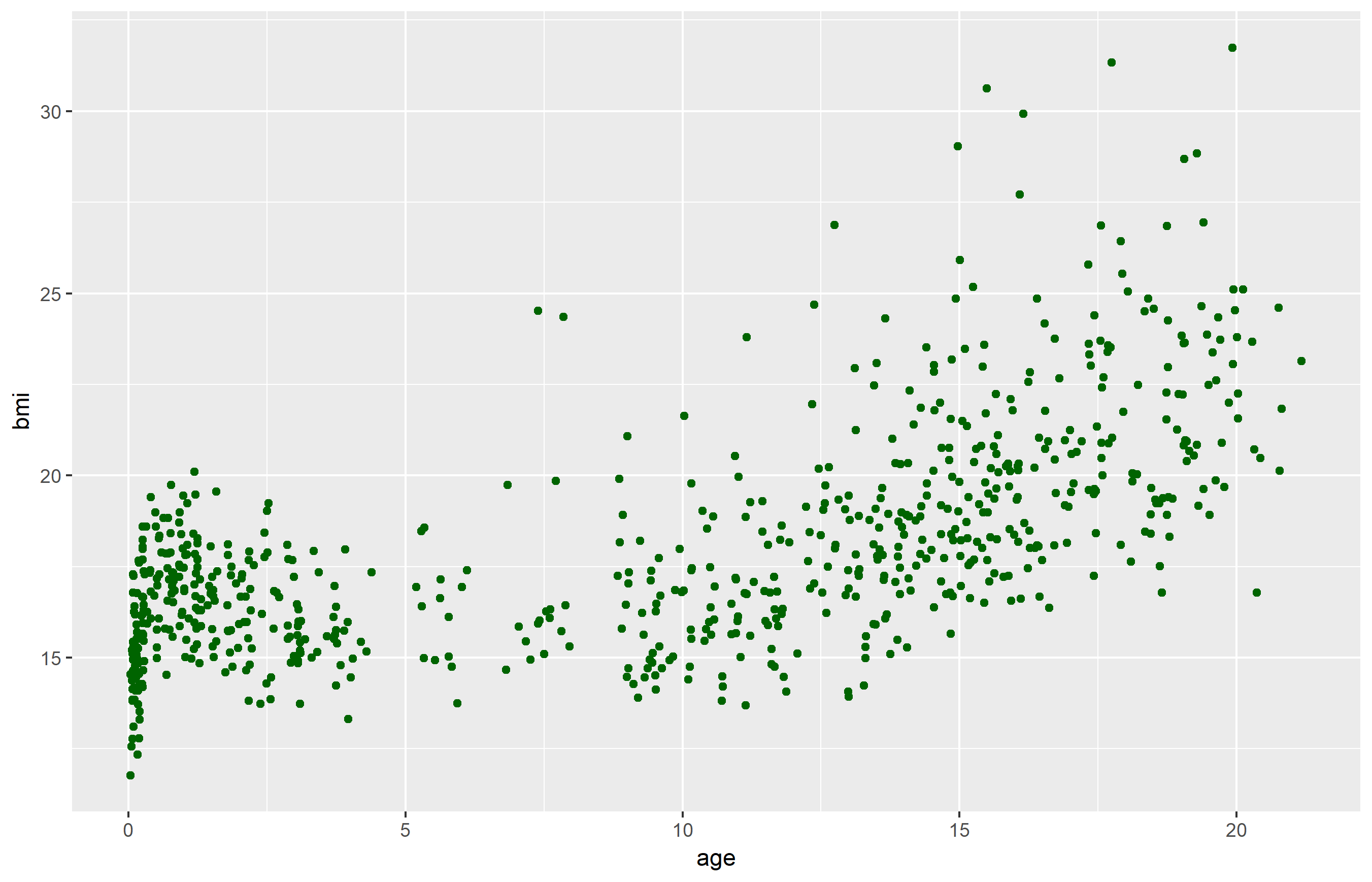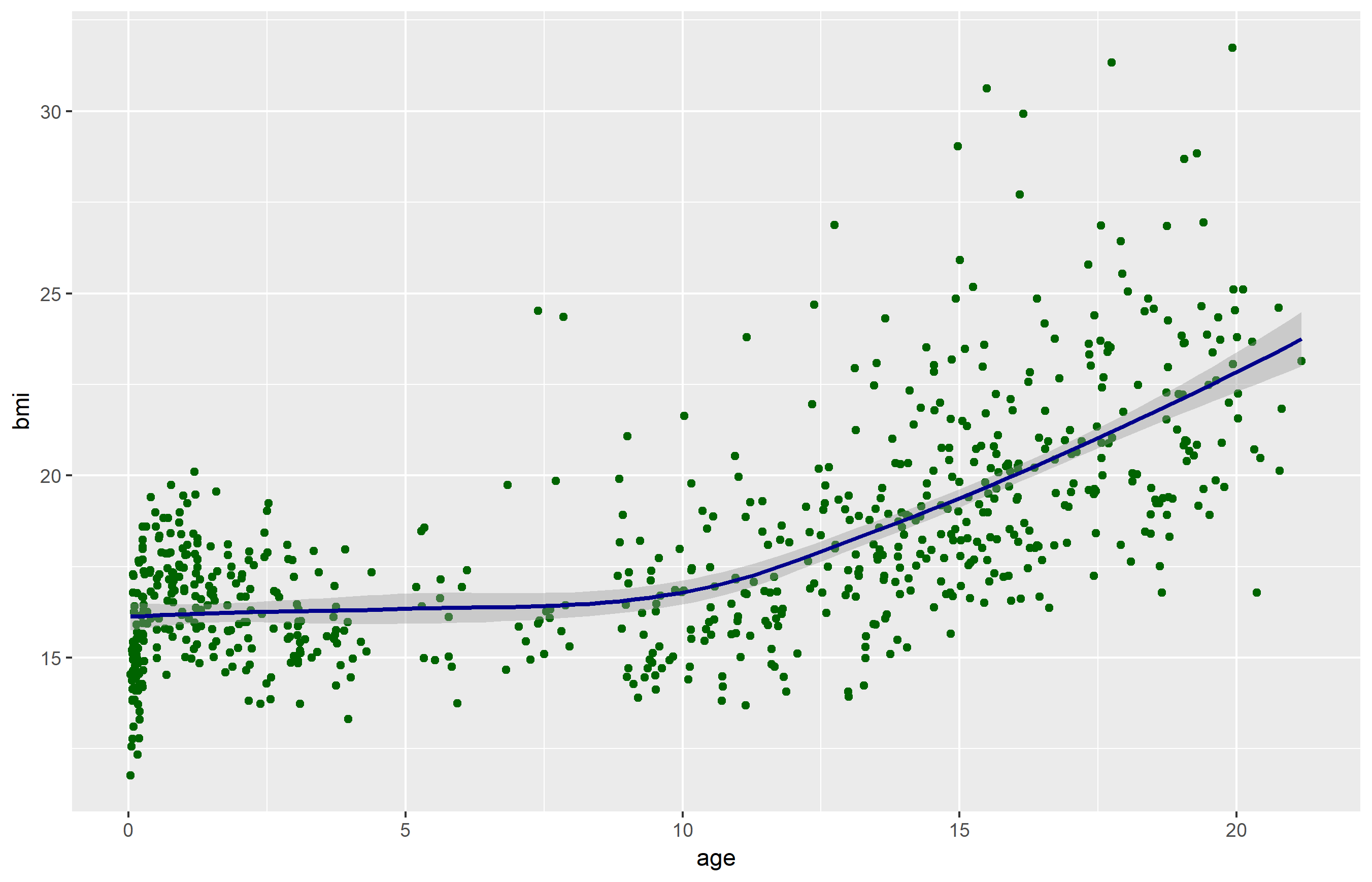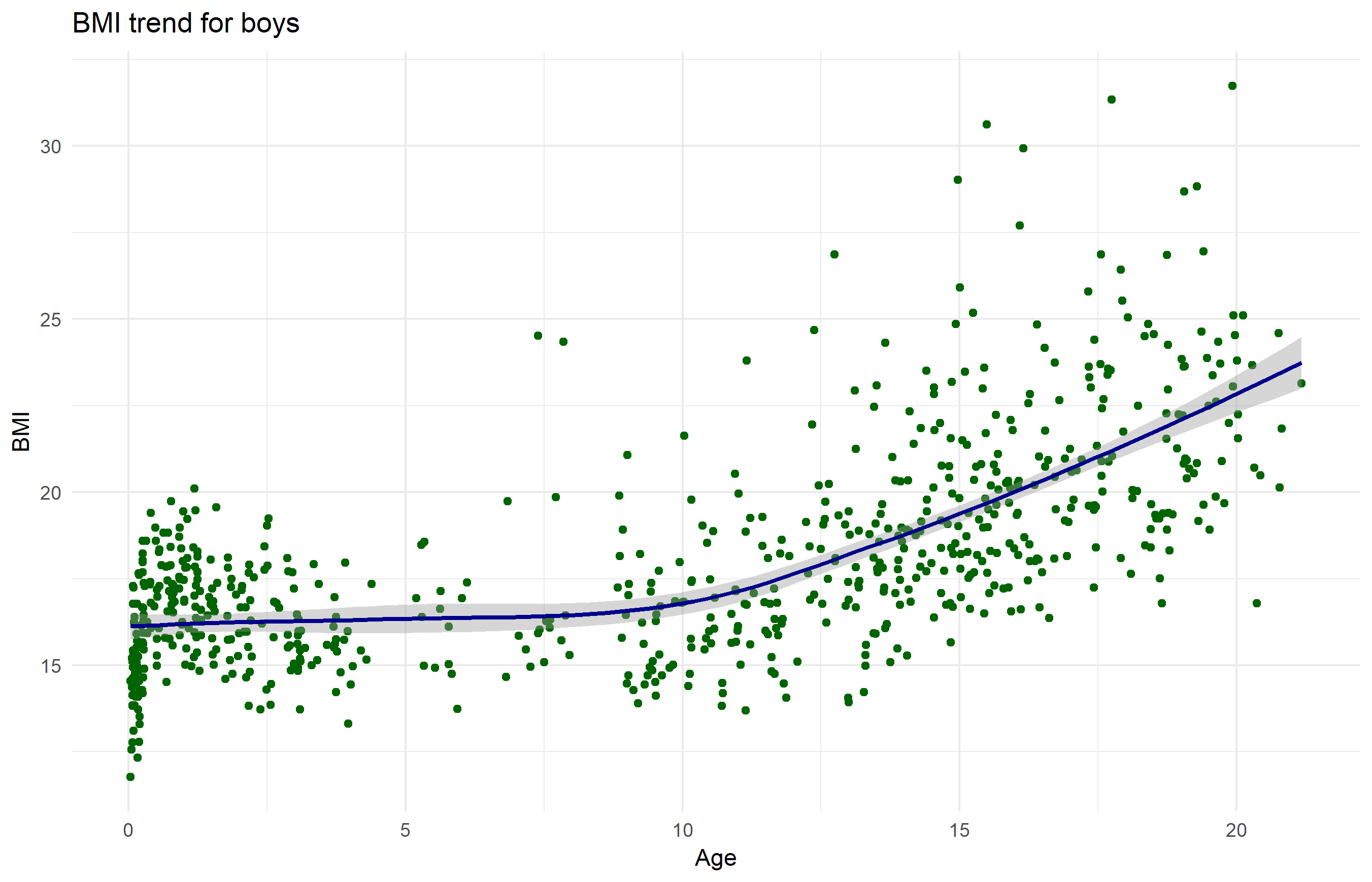
Revisit the start
Is the same as

Gerko Vink